Setup
The setup is shown schematically here:
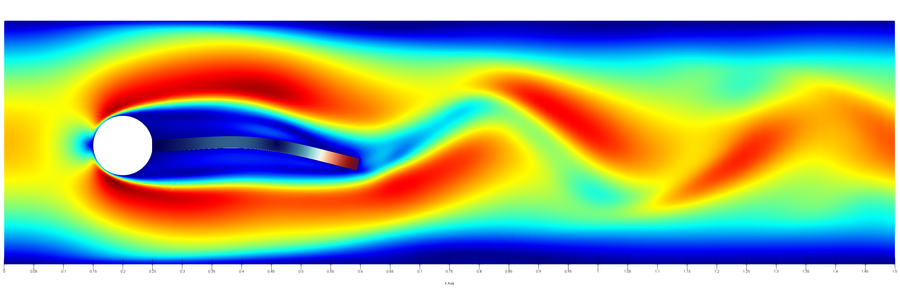
For more information please refer to the original publication of the benchmark [1].
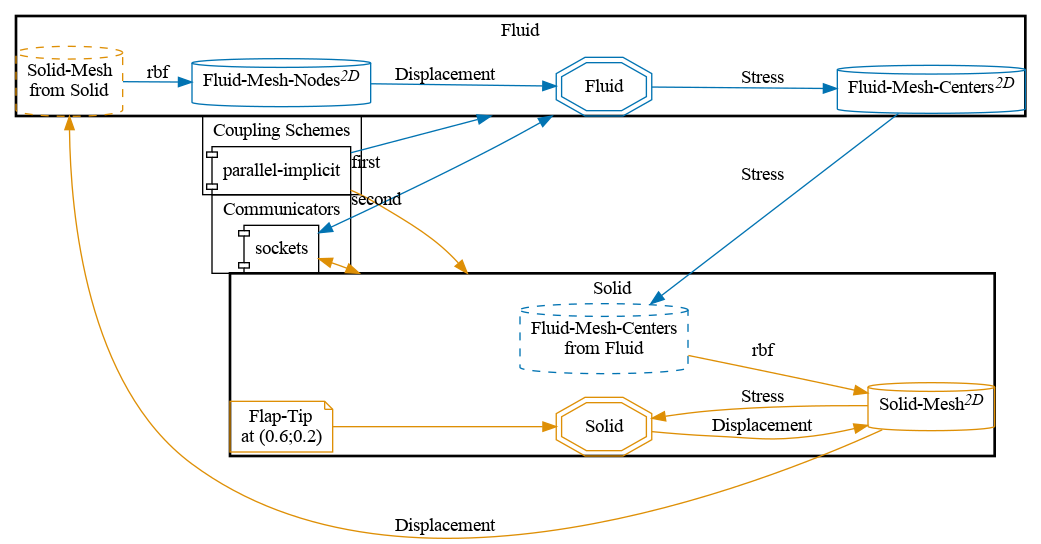
Configuration
preCICE configuration (image generated using the precice-config-visualizer):
Available solvers
Fluid participant:
- OpenFOAM (pimpleFoam). In case you are using a very old OpenFOAM version, you will need to adjust the solver to
pimpleDyMFoamin theFluid/system/controlDictfile. For more information, have a look at the OpenFOAM adapter documentation. - Nutils. For more information, have a look at the Nutils adapter documentation. This Nutils solver requires at least Nutils v9.0. This case takes significantly longer to run than the OpenFOAM case, see related issue.
Solid participant:
- deal.II. For more information, have a look at the deal.II adapter documentation. This tutorial requires the nonlinear solid solver. Please copy the nonlinear solver executable to the
solid-dealiifolder or make it discoverable at runtime and update thesolid-dealii/run.shscript. - Nutils. For more information, have a look at the Nutils adapter documentation. This Nutils solver requires at least Nutils v9.0.
Running the Simulation
Open two separate terminals and start each participant by calling the respective run script. For example:
cd fluid-openfoam
./run.sh
and
cd solid-dealii
./run.sh
You can also run OpenFOAM in parallel by ./run.sh -parallel. The default setting here uses 25 MPI ranks. You can change this setting in fluid-openfoam/system/decomposeParDict.
You may adjust the end time in the precice-config.xml, or interrupt the execution earlier if you want.
In the first few timesteps, many coupling iterations are required for convergence. Don’t lose hope, things get better quickly.
Post-processing
You can visualize the results of the coupled simulation using e.g. ParaView. OpenFOAM uses an OpenFOAM-specific format, and you can directly load the (empty) file fluid-openfoam.foam in Paraview or convert the results to VTK with foamToVTK. deal.II writes VTK files. Both Nutils solvers currently do not write VTK files (but use their own in-situ visualization), but this can be easily added similarly to the perpendicular flap solvers.
If you want to visualize both domains with ParaView, keep in mind that the different solvers may write results with different output frequencies, which you might want to synchronize.
There is a known issue that leads to additional “empty” result directories when running with some OpenFOAM versions, leading to inconveniences during post-processing. At the end of run.sh, we call openfoam_remove_empty_dirs (provided by tools/openfoam-remove-empty-dirs) to delete the additional files before importing the results in ParaView.
Moreover, as we defined a watchpoint at the flap tip (see precice-config.xml), we can plot it with gnuplot using the script plot-displacement.sh, which expects the directory of the selected solid participant as a command line argument. For example:
plot-displacement.sh solid-dealii
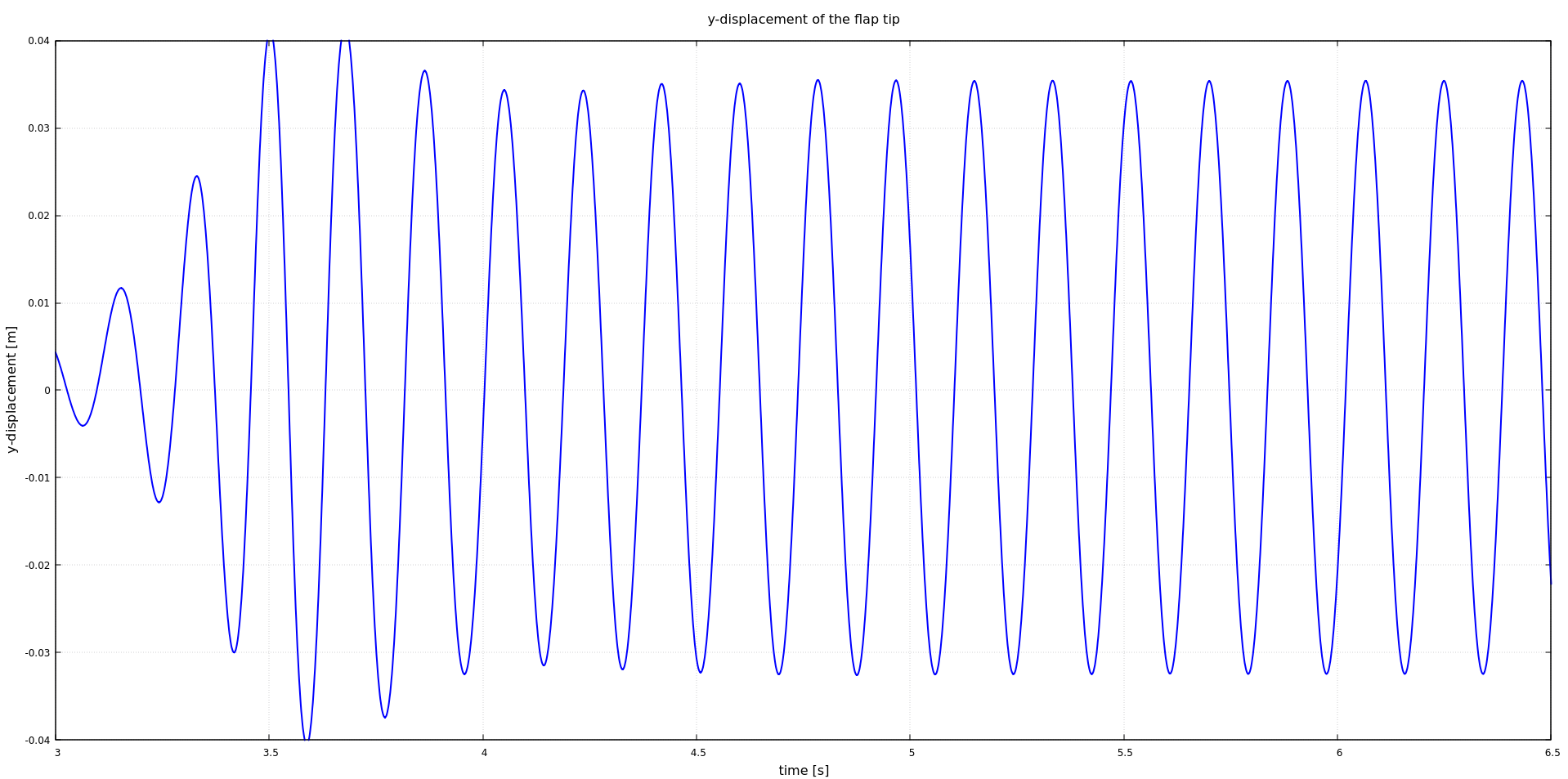
Before running the simulation again, you may want to cleanup any result files using the script `clean-tutorial.sh`.
## Mesh refinement
In `fluid-openfoam/system/`, we provide three different fluid meshes:
* `blockMeshDict`: the default mesh with approximately 21k cells,
* `blockMeshDict_refined`: a refined mesh with approximately 38k cells,
* `blockMeshDict_double_refined`: a refined mesh with approximately 46k cells.
If you want to use one of the two refined meshes, simply swap the `blockMeshDict`:
```bash
mv blockMeshDict blockMeshDict_original
mv blockMeshDict_refined blockMeshDict
Acknowledgements
Thanks to the Technical University of Eindhoven for funding the development of the Nutils participants for this tutorial.
References
[1] S. Turek, J. Hron, M. Madlik, M. Razzaq, H. Wobker, and J. Acker. Numerical simulation and benchmarking of a monolithic multigrid solver for fluid-structure interaction problems with application to hemodynamics. In H.-J. Bungartz, M. Mehl, and M. Schäfer, editors, Fluid Structure Interaction II: Modelling, Simulation, Optimization, page 432. Springer Berlin Heidelberg, 2010.